Search
About Hortaea werneckii (GCA_003704385.1)
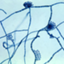
Hortaea werneckii is a species of yeast in the family Teratosphaeriaceae. Several salt-tolerance mechanisms of H. werneckii have been studied on molecular level. For example, it is known that its major compatible solutes are glycerol, erythritol, arabitol, and mannitol; melanin accumulation of the cell wall aids in retention of at least glycerol inside of the cell. Several components of the high osmolarity glycerol (HOG) signalling pathway (which controls responses to osmotic shock) have been studied in detail and some seem to differ in function compared to their counterparts in Saccharomyces cerevisiae. Whole genome sequencing of H. werneckii Genes encoding metal cation transporters, which are thought to play a role in halotolerance, experienced several additional gene duplications at various points during their evolution. A homothallic mating locus was found in all sequenced genomes, although one of the mating genes may have been inactivated in some strains. Despite this, phylogenetic analyses and linkage disequilibrium analyses indicate that H. werneckii is asexual.
(Text from Wikipedia and [image] (https://commons.wikimedia.org/wiki/File:Hortaea-werneckii-fungus--causes-tinea-nigra.jpg) from Wikipedia (http://en.wikipedia.org), the free encyclopaedia.
Taxonomy ID 91943
Data source Biotechnical Faculty, University of Ljubljana
Comparative genomics
What can I find? Homologues, gene trees, and whole genome alignments across multiple species.
 More about comparative analyses
More about comparative analyses
 Phylogenetic overview of gene families
Phylogenetic overview of gene families
 Download alignments (EMF)
Download alignments (EMF)
Variation
This species currently has no variation database. However you can process your own variants using the Variant Effect Predictor:



 {#wiki_icon}
{#wiki_icon}
 Display your data in Ensembl Fungi
Display your data in Ensembl Fungi

 Update your old Ensembl IDs
Update your old Ensembl IDs
![Follow us on Twitter! [twitter logo]](/i/twitter.png)
