Search
About Aspergillus nidulans FGSC A4
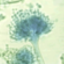
Aspergillus nidulans is one of many species of filamentous fungi in the phylum Ascomycota. It has been an important research organism for studying eukaryotic cell biology
for over 50 years,
being used to study a wide range of subjects including recombination, DNA repair, mutation, cell cycle control, tubulin, chromatin, nucleokinesis, pathogenesis, metabolism, and experimental evolution.
It is one of the few species in its genus able to form sexual spores through meiosis, allowing crossing of strains in the laboratory. A. nidulans is a homothallic fungus, meaning it is able to self-fertilize and form fruiting bodies in the absence of a mating partner. It has septate hyphae with a woolly colony texture and white mycelia. The green colour of wild-type colonies is due to pigmentation of the spores, while mutations in the pigmentation pathway can produce other spore colours.
Picture credit: Creative Commons Attribution-Share Alike 3.0 via Wikimedia Commons (Image source) Taxonomy ID 227321
(Text from Wikipedia.)
More information General information about this species can be found in Wikipedia
Taxonomy ID 227321
Data source European Nucleotide Archive
Comparative genomics
What can I find? Homologues, gene trees, and whole genome alignments across multiple species.
 More about comparative analyses
More about comparative analyses
 Phylogenetic overview of gene families
Phylogenetic overview of gene families
 Download alignments (EMF)
Download alignments (EMF)
Variation
This species currently has no variation database. However you can process your own variants using the Variant Effect Predictor:




 Display your data in Ensembl Fungi
Display your data in Ensembl Fungi

 Update your old Ensembl IDs
Update your old Ensembl IDs

![Follow us on Twitter! [twitter logo]](/i/twitter.png)
