SuperContig FN430008: 277,519-279,869 reverse strand.
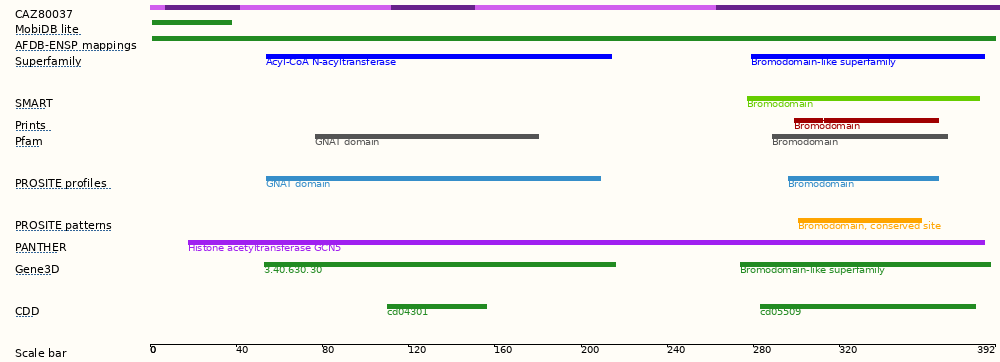
This transcript has 6 exons and is annotated with 20 domains and features.
This transcript is a product of gene GSTUM_00001663001 Show transcript tableHide transcript table
 Tuber melanosporum (ASM15164v1) ▼
Tuber melanosporum (ASM15164v1) ▼  Summary
Summary Sequence
Sequence Protein Information
Protein Information Protein summary
Protein summary Domains & features
Domains & features Variants
Variants PDB 3D protein model
PDB 3D protein model AlphaFold predicted model
AlphaFold predicted model Genetic Variation
Genetic Variation Variant table
Variant table Variant image
Variant image Population comparison
Population comparison Comparison image
Comparison image External References
External References General identifiers
General identifiers Oligo probes
Oligo probes Supporting evidence
Supporting evidence ID History
ID History.
.
SuperContig FN430008: 277,519-279,869 reverse strand.
This transcript has 6 exons and is annotated with 20 domains and features.
This transcript is a product of gene GSTUM_00001663001 Show transcript tableHide transcript table
| Name | Transcript ID | bp | Protein | Biotype | UniProt | RefSeq | Flags |
|---|---|---|---|---|---|---|---|
| GSTUM_00001663001/CAZ80037 | CAZ80037 | 1702 | 392aa | Gene/transcipt that contains an open reading frame (ORF).Protein coding | D5G694 | - | A single transcript chosen for a gene which is the most conserved, most highly expressed, has the longest coding sequence and is represented in other key resources, such as NCBI and UniProt. This is defined in detail on http://www.ensembl.org/info/genome/genebuild/canonical.htmlEnsembl Canonical, |
.
Cleavage site (Signalp)
Signal peptide cleavage sites predicted by SignalP.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Low complexity (Seg)
Low complexity peptide sequences identified by Seg.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Transmembrane helices
Transmembrane helices predicted by TMHMM.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
HAMAP
HAMAP families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
NCBIFAM
NCBIfam is a collection of protein families based on Hidden Markov Models (HMMs).
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PIRSF
Protein domains and motifs from the PIR (Protein Information Resource) Superfamily database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
SFLD
Structure-Function Linkage Database families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Coiled-coils (Ncoils)
Coiled-coil regions predicted by Ncoils.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
TIGRFAM
Protein domains and motifs from the TIGRFAM database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
MobiDB lite
Intrinsically disordered regions predicted by MobiDB lite.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
AFDB-ENSP mappings
Protein features that represent the mapping between Ensembl proteins (ENSP) and AlphaFoldDB protein structures (including their corresponding chains). Imported via UniProt mappings
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Superfamily
Protein domains and motifs from the SUPERFAMILY database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
SMART
Protein domains and motifs from the SMART database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Prints
Protein fingerprints (groups of conserved motifs) from the PRINTS database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Pfam
Protein domains and motifs from the Pfam database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PROSITE profiles
Protein domains and motifs from the PROSITE profiles database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PROSITE patterns
Protein domains and motifs from the PROSITE patterns database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
PANTHER
PANTHER families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Gene3D
CATH/Gene3D families.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
CDD
Conserved Domain Database models.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Ave. residue weight: 114.928 g/mol
Charge: 17.0
Isoelectric point: 9.5866
Molecular weight: 45,051.61 g/mol
Number of residues: 392 aa
.
Ensembl Fungi release 60 - October 2024 © EMBL-EBI EMBL-EBI
.
.
This website requires cookies, and the limited processing of your personal data in order to function. By using the site you are agreeing to this as outlined in our Privacy Policy and Terms of Use