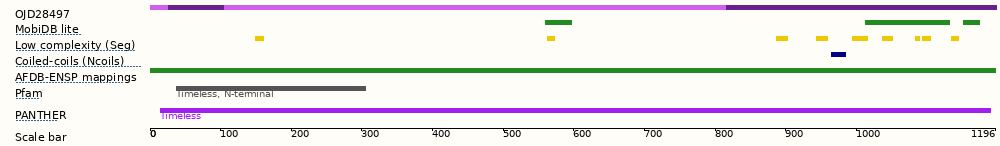
[["rect",[150,5,170,10],{"title":"Exon: OJD28497-1; Location: <a href=\"/Blastomyces_percursus_gca_001883805/Location/View?db=core;g=ACJ73_00096;r=supercont2.5:28690-32482;t=OJD28497\">supercont2.5:28690-32482</a>; First aa: 1 ; Last aa: 26 ; Start phase: 0; End phase: 2; Length: 78bp, 26 aa"}],["rect",[168,5,225,10],{"title":"Exon: OJD28497-2; Location: <a href=\"/Blastomyces_percursus_gca_001883805/Location/View?db=core;g=ACJ73_00096;r=supercont2.5:28690-32482;t=OJD28497\">supercont2.5:28690-32482</a>; First aa: 27 ; Last aa: 106 (1st base); Start phase: 0; End phase: 0; Length: 238bp, 79 1/3 aa"}],["rect",[224,5,728,10],{"title":"Exon: OJD28497-3; Location: <a href=\"/Blastomyces_percursus_gca_001883805/Location/View?db=core;g=ACJ73_00096;r=supercont2.5:28690-32482;t=OJD28497\">supercont2.5:28690-32482</a>; First aa: 106 (2nd base); Last aa: 817 (1st base); Start phase: 1; End phase: 0; Length: 2133bp, 711 aa"}],["rect",[726,5,997,10],{"title":"Exon: OJD28497-4; Location: <a href=\"/Blastomyces_percursus_gca_001883805/Location/View?db=core;g=ACJ73_00096;r=supercont2.5:28690-32482;t=OJD28497\">supercont2.5:28690-32482</a>; First aa: 817 (2nd base); Last aa: 1197 ; Start phase: 1; End phase: 2; Length: 1142bp, 380 2/3 aa"}],["rect",[15,5,64,19],{"klass":["label","DmWYEpiV"],"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)"}],["rect",[545,20,572,25],{"title":"MobiDB lite; Position: 560-596"}],["rect",[865,20,950,25],{"title":"MobiDB lite; Position: 1012-1131"}],["rect",[889,20,911,25],{"title":"MobiDB lite; Position: 1046-1077"}],["rect",[922,20,949,25],{"title":"MobiDB lite; Position: 1094-1130"}],["rect",[963,20,980,25],{"title":"MobiDB lite; Position: 1152-1174"}],["rect",[15,20,82,34],{"klass":["label","CXukBbYA"],"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)"}],["rect",[255,36,264,41],{"title":"Low complexity (Seg); Position: 149-161"}],["rect",[547,36,555,41],{"title":"Low complexity (Seg); Position: 562-572"}],["rect",[776,36,788,41],{"title":"Low complexity (Seg); Position: 887-902"}],["rect",[816,36,828,41],{"title":"Low complexity (Seg); Position: 943-959"}],["rect",[852,36,868,41],{"title":"Low complexity (Seg); Position: 995-1016"}],["rect",[882,36,893,41],{"title":"Low complexity (Seg); Position: 1037-1051"}],["rect",[915,36,920,41],{"title":"Low complexity (Seg); Position: 1083-1089"}],["rect",[922,36,931,41],{"title":"Low complexity (Seg); Position: 1094-1105"}],["rect",[951,36,959,41],{"title":"Low complexity (Seg); Position: 1134-1145"}],["rect",[15,36,136,50],{"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)","klass":["label","0I24elt1"]}],["rect",[831,52,846,57],{"title":"Coiled-coils (Ncoils); Position: 964-984"}],["rect",[15,52,142,66],{"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)","klass":["label","5lAVRiHi"]}],["rect",[150,68,996,73],{"title":"AFDB-ENSP mappings; Position: 1-1196"}],["rect",[15,68,124,82],{"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)","klass":["label","o3WxLZKC"]}],["rect",[176,84,366,98],{"title":"Pfam PF04821; Positions: 37-305; Interpro: IPR006906; Timeless, N-terminal","href":"/Blastomyces_percursus_gca_001883805/ZMenu/Transcript/ProteinSummary?config=protview;db=core;g=ACJ73_00096;pf_id=65090955;r=supercont2.5:28690-32482;t=OJD28497;track=domain_pfam;translation_id=1784351"}],["rect",[15,84,40,98],{"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)","klass":["label","fcCyNC0R"]}],["rect",[160,106,991,120],{"title":"PANTHER PTHR22940; Positions: 15-1190; Interpro: IPR044998; Timeless","href":"/Blastomyces_percursus_gca_001883805/ZMenu/Transcript/ProteinSummary?config=protview;db=core;g=ACJ73_00096;pf_id=65090962;r=supercont2.5:28690-32482;t=OJD28497;track=domain_hmmpanther;translation_id=1784351"}],["rect",[15,106,58,120],{"klass":["label","tz3TYWb3"],"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)"}],["rect",[15,128,70,142],{"klass":["label","DypjnZKj"],"alt":"Blastomyces percursus str. EI222 (Blastomyces percursus str. EI222)"}]]
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE profiles database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein fingerprints (groups of conserved motifs) from the PRINTS database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
NCBIfam is a collection of protein families based on Hidden Markov Models (HMMs).
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SUPERFAMILY database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the PROSITE patterns database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the SMART database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track
Protein domains and motifs from the TIGRFAM database.
URL to turn this track on
Copy the above url to force this track to be turned on
Click on the star to add/remove this track from your favourites
Click on the cross to turn the track off
Click to highlight/unhighlight this track





![Follow us on Twitter! [twitter logo]](/i/twitter.png)
